import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from matplotlib_venn import venn3_unweighted as venn3
Extracting data¶
from google.colab import drive
drive.mount('/content/gdrive', force_remount=True)
root_dir = "/content/gdrive/My Drive/idp-ego/GUI/algorithms/trio/"
data_dir = root_dir + 'data/'
# path = Path(base_dir + 'data/bears')
# dest.mkdir(parents=True, exist_ok=True) I want only existing folders
e1=pd.read_csv(data_dir+'COL1538-V4-2.csv',low_memory=0)
e1.head(5)
e1[e1['chr']=='1'].head(5) #from 0 to 5-1
u=pd.DataFrame({'chr':['1','x','y'],'value':['val1','val2','val3']}) #example of a df creation
u
chr_e2=['1','2']
e2_filter=[x in chr_e2 for x in tuple(e1['chr'])] #search for list comprehension vs generator
#print(e2_filter)
e2=e1[e2_filter]
e2.tail(5)
chr_e3=['1','12']
e3_filter=[x in chr_e3 for x in tuple(e1['chr'])] #generator list generator
#print(e3_filter)
e3=e1[e3_filter]
print(e3.columns)
e3.tail(5)
Subsets combinations¶
available sets: e1,e2,e3 (exoms)
possible subsets:
- ${e_{_{i}}}\cap\ {e_{_{j}}}\cap\ {e_{_{k}}} \ |_{i,j,k\in \{A,B,C\}\ \wedge\ i\neq j\neq k}$
- ${e_{i}}\cap\ {e_{j}}\cap \ {{e^c}_{k}} \ |_{i,j,k\in \{A,B,C\}\ \wedge\ i\neq j\neq k}$
- ${e_{i}}\cap \ {{e^c}_{j}} \cap \ {{e^c}_{k}} \ |_{i,j,k\in \{A,B,C\}\ \wedge\ i\neq j\neq k}$
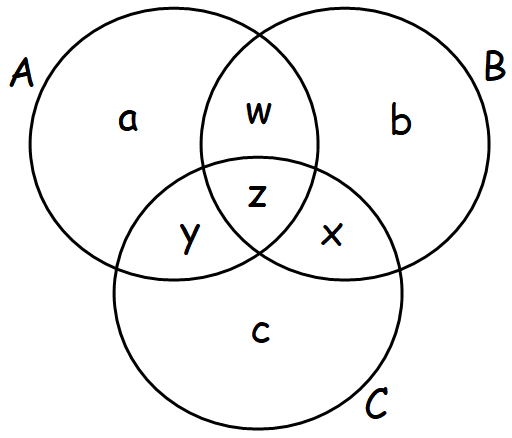
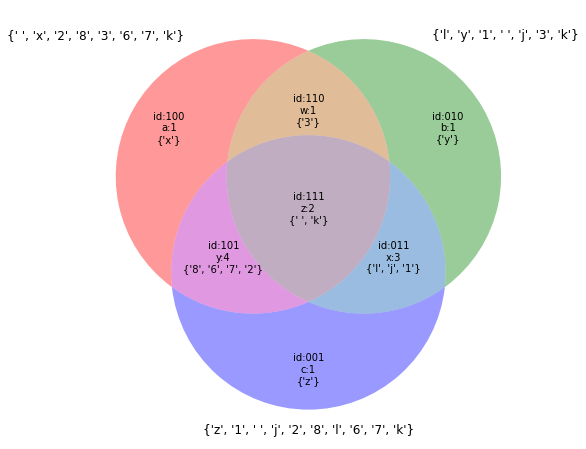
Figure 1. Venn diagram with C as the pacient, A as affected parent and B as the unaffected parent.
Requirements:
- y
- must match a triplet of (pos, ref nt, gene)
apriori_common=len(set(list(e1[e1['chr']=='1']['gene'])))
apriori_unique_e2=len(set(list(e2[e2['chr']=='2']['gene'])))
apriori_unique_e3=len(set(list(e3[e3['chr']=='12']['gene'])))
print("len(apriori_common)={}".format(apriori_common))
print("len(apriori_unique_e2)={}".format(apriori_unique_e2))
print("len(apriori_unique_e3)={}".format(apriori_unique_e3))
genes_e1=set(list(e1['gene']))
genes_e2=set(list(e2['gene']))
genes_e3=set(list(e3['gene']))
def searchforgene(gene,genes_set):
positions=[]
genes=[]
for i in range(len(genes_set)):
if gene==genes_set[i]:
positions.append(i)
genes.append(genes_set[i])
return (positions,genes)
# test
print(list(e2['gene'])[:20])
print(len(list(e2['gene'])))
print(searchforgene('DDX11L1',list(genes_e2))[0])
print(searchforgene('DDX11L1',list(genes_e2))[1])
print(searchforgene('DDX11L1',list(e2['gene']))[0])
common=genes_e1.intersection(genes_e2).intersection(genes_e3)
len(common)
The result shown above, shows that len(a_priori_common) is equal to the z subset (see fig. 1).
Constructing the generic function¶
Selecting individual tuples from the exomes files : extract_tuples_pro function¶
def extract_tuples_pro(e,prop=("gene","pos","refNt")):
"""
Must extract the desired parameters from the exome "e"
in a list of tuples(default triples)
"""
return e[list(prop)].values
def extract_tuples(e,prop=("gene","pos","refNt")):
"""
Must extract the desired parameters from the exome "e"
in a list of tuples(default triples)
"""
r={}
for i in range(len(prop)):
r[prop[i]]=tuple(e[prop[i]])
# rr[0]=("a_gen","a_pos","a_refNt")
rr=[]
for i in range(len(r[prop[0]])):
rri=[r[j][i] for j in prop]#list comprehension
rr.append(rri)
return r,rr
# test
print(extract_tuples(e2)[1][:5])
print(len(extract_tuples(e2)[0]["gene"]))
print(len(extract_tuples(e2)[0]["pos"]))
print(len(extract_tuples(e2)[0]["refNt"]))
Splitting by subsets : subset function¶
def subset(Ax,Bx,Cx,prop=("gene","pos","refNt")):
"""
Must obtain the different subsets to contruct the Venn
diagram.
A and B: parents
C: Child
"""
A=frozenset(tuple(m) for m in extract_tuples_pro(Ax,prop)) #as set 1
B=frozenset(tuple(m) for m in extract_tuples_pro(Bx,prop)) #as set 2
C=frozenset(tuple(m) for m in extract_tuples_pro(Cx,prop)) #as set 3
a=((A-B)-C) #001
b=((B-A)-C) #010
c=((C-B)-A) #100
y=((A&C)-B) #101
w=((A&B)-C) #011
x=((B&C)-A) #110
z=A.intersection(B,C) #111
combinations=[a,b,w,c,y,x,z]
merge_vector=[bin(i)[2:].zfill(3) for i in range (1,2**3)]
r=dict(zip(merge_vector,combinations))
return r
# debug
merge_vector=[bin(i)[2:].zfill(3) for i in range (1,2**3)]
print(merge_vector)
Venn Diagram¶
# Venn Diagramm - ilustrative example
A,B,C=set('23 xk678'),set('13 ykjl'),set('12 zkjl678')
Vtags=[A,B,C]
a=((A-B)-C) #001
b=((B-A)-C) #010
c=((C-B)-A) #100
y=((A&C)-B) #101
w=((A&B)-C) #011
x=((B&C)-A) #110
z=A.intersection(B,C) #111
combinations=[len(i) for i in [a,b,w,c,y,x,z]]
print(combinations)
merge_vector=[bin(i)[2:].zfill(3) for i in range (1,2**3)]
r=dict(zip(merge_vector,combinations))
T=[a,b,w,c,y,x,z]
ñ=dict(zip(merge_vector,T))
print(r)
print(merge_vector)
#(Abc, aBc, ABc, abC, AbC, aBC, ABC)
tags=['a','b','w','c','y','x','z']
q=dict(zip(merge_vector,tags))
plt.figure(figsize=(10,8))
v=venn3(combinations,Vtags)
for i in merge_vector:
try:
t=v.get_label_by_id(i).get_text()
v.get_label_by_id(i).set_text(f"id:{i}\n{q[i[::-1]]}:{t}\n{ñ[i[::-1]]}")
except:
print("debuging bro, testing")
plt.show()
Venn Function¶
def venn(r,Vtags=('A','B','C')):
plt.figure(figsize=(10,8))
#(Abc, aBc, ABc, abC, AbC, aBC, ABC) <- format of venn3
# str[::-1] is used to flip strings
mv=[bin(i)[2:].zfill(3) for i in range (1,2**3)]
tags=['a','b','w','c','y','x','z']
q=dict(zip(mv,tags))
v=venn3([len(r[i]) for i in mv],Vtags)
for i in mv:
t=v.get_label_by_id(i).get_text()
v.get_label_by_id(i).set_text(f"id:{i}\n{q[i[::-1]]}\n{t}")
plt.show()
return None
Implementing and testing the function with sets¶
# a_priori test
prop=("gene","pos","refNt")
apriori_common=len(e1[list(prop)])
print("len(apriori_common)={}".format(apriori_common))
print(e1[list(prop)].values[:5])
e1[list(prop)].head(5)
A=frozenset(tuple(u) for u in extract_tuples(e1)[1])
B=frozenset(tuple(u) for u in extract_tuples(e2)[1])
C=frozenset(tuple(u) for u in extract_tuples(e3)[1])
z=A.intersection(B,C)
print(len(z))
y=len((A&C)-B)
print(y)
# proving the triplets actually match in the analysed case (z)
aux1=e1[["chr","gene","pos","refNt"]]
print(type(aux1))
aux1=aux1[aux1['chr']=='1']
print(type(aux1))
print(len(aux1))
aux1=aux1[["gene","pos","refNt"]].values
aux1=frozenset(tuple(u) for u in aux1)
print(len(aux1))
Time test¶
# time test
%timeit -n 10 -r 5 extract_tuples(e1)
%timeit -n 10 -r 5 extract_tuples_pro(e1)
As a conclusion, the function extract_list_pro spent about 10% of the time spent by the extract_tuples function
Extending to the real case¶
e1=pd.read_csv(data_dir+'COL1541-V4-2.csv',low_memory=0) #father
e2=pd.read_csv(data_dir+'COL1539-V4-2.csv',low_memory=0) #mother
e3=pd.read_csv(data_dir+'COL1538-V4-2.csv',low_memory=0) #child
Example 1: prop=("gene","pos","refNt") triplet¶
There is no need to call prop=("gene","pos","refNt") as this is a default argument of the subset function.
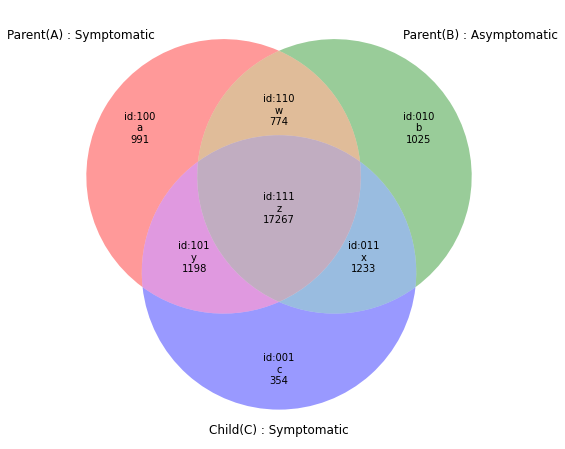
r=subset(e1,e2,e3)
venn(r,Vtags=("Parent(A) : Symptomatic","Parent(B) : Asymptomatic","Child(C) : Symptomatic"))#217997
Data$\rightarrow$DataFrame¶
Because each subset is formed of unique tuples the DataFrame constructor must be used (then if desired, convert it to excel. Also to alocate the desired subset, the id displayed in the section Example 1 must be used as a key to access every element of the dictionary.
prop=("gene","pos","refNt")
pd.DataFrame(r["111"],columns=prop).head(5)
Testing¶
print(f"len(r['101'])={len(r['101'])}")
print(f"len(r['101']&r['001'])={len(r['100'].intersection(r['001']))}")
Note that len(r['101']&r['001'])=0 makes sense due to the fact that the subsets are already separed
# testing subset function with Venn diagram 'created' from scratch
At=frozenset(tuple(m) for m in extract_tuples_pro(e1))
Bt=frozenset(tuple(m) for m in extract_tuples_pro(e2))
Ct=frozenset(tuple(m) for m in extract_tuples_pro(e3))
a=((At.difference(Bt)).difference(Ct)) #001
b=((Bt.difference(At)).difference(Ct)) #010
c=((Ct.difference(Bt)).difference(At)) #100
y=((At.intersection(Ct)).difference(Bt)) #101
w=((At.intersection(Bt)).difference(Ct)) #011
x=((Bt.intersection(Ct)).difference(At)) #110
z=At.intersection(Bt,Ct) #111
venn3(subsets=[len(i) for i in [a,b,w,c,y,x,z]])
Example 2: Custom prop parameter¶
Setting prop=("gene") will establish a different filter for the analysis
r2=subset(e1,e2,e3,prop=("gene",))
venn(r2,Vtags=("Parent(A) : Symptomatic","Parent(B) : Asymptomatic","Child(C) : Symptomatic"))
Example 3: Custom prop parameter¶
Setting prop=("pos") will establish a different filter for the analysis
r3=subset(e1,e2,e3,prop=("pos",))
venn(r3,Vtags=("Parent(A) : Symptomatic","Parent(B) : Asymptomatic","Child(C) : Symptomatic"))#217906
Example 4: Custom prop parameter¶
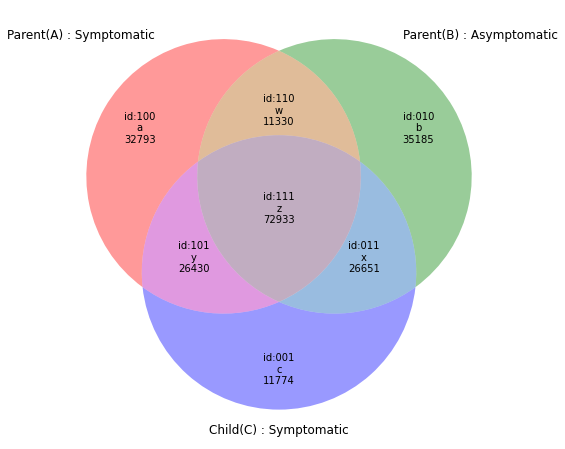
r4=subset(e1,e2,e3,prop=("gene","pos","refNt","altNt"))
venn(r4,Vtags=("Parent(A) : Symptomatic","Parent(B) : Asymptomatic","Child(C) : Symptomatic"))#218853
Data $\rightarrow$ DataFrame¶
prop2=("gene","pos","refNt","altNt")
pato=pd.DataFrame(r4["101"],columns=prop2).head(5)
pato
pato.to_csv()
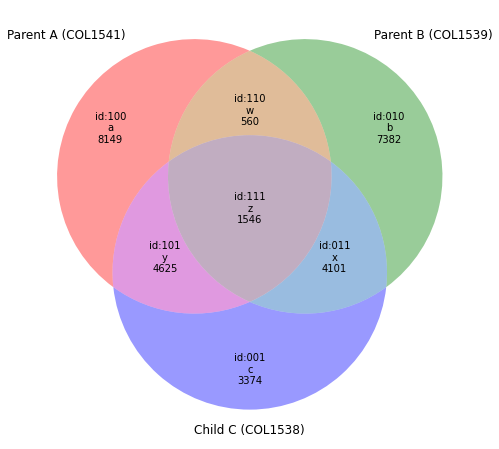
Application: Comparison between exomes analysis¶
#loading files
A=pd.read_csv(root_dir+'data-us/COL1541.V4.2.gnomGN_0.05.csv',low_memory=0) #father
B=pd.read_csv(root_dir+'data-us/COL1539.V4.2.gnomGN_0.05.csv',low_memory=0) #mother
C=pd.read_csv(root_dir+'data-us/COL1538.V4.2.gnomGN_0.05.csv',low_memory=0) #child
#plotting venn
prop_=("gene","pos","refNt","altNt")#illustrative
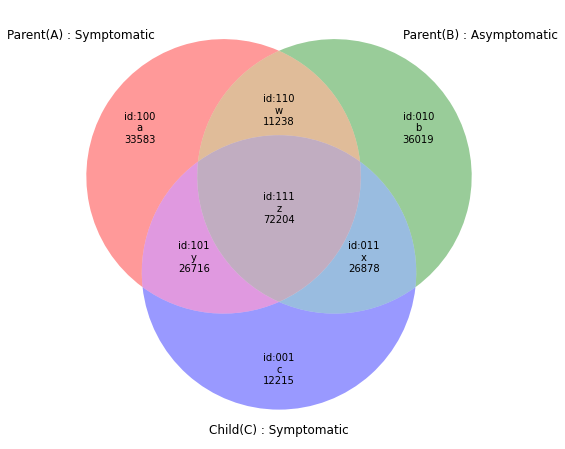
QQ=subset(A,B,C,prop=prop_)
venn(QQ,Vtags=("Parent A (COL1541)","Parent B (COL1539)","Child C (COL1538)"))
Data $\rightarrow$ DataFrame¶
duck=pd.DataFrame(QQ["011"],columns=prop_).head(10)
duck